From cancer prediction to rare disease diagnosis...World's Largest Analysis of Genetic Function
|
Professor Kim Hyung-beom, lecturer Lee Kwang-seop, and graduate student research team at Yonsei University Medical School announced on the 15th that they succeeded in evaluating 27513 single base mutations of ATM genes closely related to cancer and rare diseases.
The results of the study were published in the latest issue of the international journal Cell (IF 42.5).
ATM genes play an important role in detecting and repairing DNA damage in the body. If this gene does not work properly, there is a high risk of cancer such as breast cancer, colon cancer, and pancreatic cancer, and the prognosis of cancer patients is often poor. It can also cause certain rare diseases, such as 'exercise-capillary dilatation'. Discovering mutations that disrupt the function of the ATM gene can predict the risk of developing cancer and the prognosis of treatment for cancer patients in healthy general populations with the mutation.
With the recent development of dielectric analysis technology, genetic disease and cancer diagnosis have become more precise, but many genetic mutations are still not properly used for treatment and diagnosis of patients because it is not known whether they are harmful or not. In particular, the ATM gene is a large gene with about 9000 protein base sequences and has a large number of mutations, making it difficult to evaluate with existing statistical methods, making it difficult to use for actual patient treatment and diagnosis.
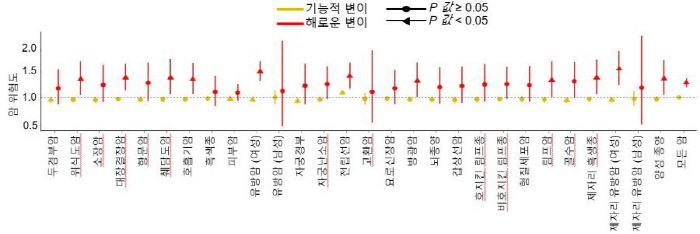
The research team analyzed 27,513 single base mutations that could occur at the entire protein coding site (62 exons) of the ATM gene. Of these, 23,092 variations were directly confirmed by cell experiments using the latest gene editing technology 'prime editing', and the remaining 4,421 variations, which are difficult to evaluate experimentally, were predicted to have an impact on cell survival using their own artificial intelligence model 'DeepATM'.
Analysis of the effect of each variation on cell survival allowed us to distinguish between variants that were detrimental to the function of the corresponding gene and those that were not with high accuracy.
The research team verified the genome and clinical data of about 500,000 people in the UK Biobank and confirmed that people with harmful mutations identified in the study have a 1.4 times higher risk of cancer than those who do not. We also demonstrated that the results matched more than 95% with data from ClinVar, an international genetic mutation database. The results of reinterpreting existing cancer genomic data (cBioPortal) from the perspective of this study also confirmed that the survival rate of cancer patients varies depending on whether ATMs have harmful mutations.
Professor Kim Hyung-beom said, `This study is of great significance in that it provides the possibility to accurately identify mutations in ATM genes that are difficult to interpret on a large scale.' `In the future, similar analysis will be possible in other genes, and this will contribute to the development of precision medicine based on dielectric substance.'
|
This article was translated by Naver AI translator.